The idea
Maybe you still know it from school: Each cell of a human being has two sets of 23 chromosomes, each set received from one parent. Every chromosome is a long and continuous piece of DNA (Deoxyribonucleic acid) which contains many genes and other elements. The backbone of the DNA carries four types of molecules called bases (A, C, G, T). The sequence of these four bases encodes the information and is the genetic code of all of us (more information…).
We took the genetic code from huge data files and assigned a color to every of the four bases. Then we rendered these fascinating pictures displaying each base by one pixel, showing the genetic code of humans in color. The resulting images have a width of 3,500 pixels and a height over 70,000 pixels for the largest chromosomes.
Although scientists know already most of the sequence of the human DNA, the information the genetic code holds is not fully understood. Therefore we want to introduce another idea to display and maybe better understand the code. Structures and repeating bases can be recognized faster and by resizing an image with an appropriate algorithm it is possible to get a quick overview of different sections in the chromosome.
FAQs
Why are there gray lines and areas in the DNA pictures?
- By far not all bases of the chromosomes are known. There are quite a few places where it is very difficult or impossible to determine the bases with today known equipment. Therefore the gray pixels and lines indicate unknown areas of the chromosome.
Why are there slight structures in the images?
- Regular pattern (like diagonal stripes) can be seen very easily and result from identical base sequences, which repeat after a constant number of bases. But you can also see slight structures and strange patterns in the pictures which are spread all over the whole chromosomes (best viewed when shutting your eyes just a little bit. Click on a link to a chromosome above and use your imagination to get a new view of your genes).
- These arcs and circles might result from identical or similar base sequences that repeat after a variable number of bases. The appearance of the arcs does not seem to correlate to any obvious genetic context (e.g. to gene-rich/ gene-poor chromosomal regions, content of A/T or G/C bases in the sequence). Maybe there is additional information modulated into the chromosome (comparable to FM radio where a lowpass signal (information 1) is transmitted over a bandpass channel (information 2)). This is open to discussion and we would appreciate comments and ideas.
- It is important to note that the structures depend on the image width. If an image is rendered with a different width, you get a different structure. But it seems that the structures occur on most sizes.
Can the slight structures be extracted out of the images to enhance their visibility?
- There are several ways to extract low frequency pattern from noisy images. For example one may just use a low frequency filter to enhance the visibility of the slight structure somewhat. But we tried a very interesting mathematical method called stochastic resonance (SR), which leads to a visual perception of a sharpened pattern by adding noise to the noisy image and thus imitating the visual perception when watching the pictures with squeezed eyes. How this works is described in Pattern & SR.
What did you decide when rendering the chromosomal pictures? What problems did you encounter?
- We decided what color each base has. The picture would look different of course when we would have selected other colors, but we went close to colors used when determining DNA sequences. Furthermore we decided to set the width of every image to 3,500 pixels.
- We had to cope with single data file sizes with more than 250 Megabytes and ended with an image height of over 70,000 pixels only for chromosome 1. Rendering all chromosomes takes hours.
What happens when the bases of the DNA are colored pairwise?
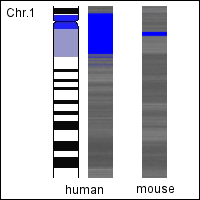
- When the bases are colored pairwise e.g. adenine/thymine in black and cytosine/guanine in white, the resulting chromosomal images obtain a characteristic banding pattern similar to the banding pattern of experimentally stained chromosomes. We compared these rendered human chromosomes with the schematic representation of the experimentally stained chromosomes (Ideograms) and also with other species (Species comparison) finding interesting correlations.
There exists also extrachromosomal DNA. Can it be rendered, too?
-
- Apart from our chromosomes we have also genetic material in cell organells called mitochondria. This mitochondrial DNA (mtDNA) is a circular molecule and much smaller than chromosomal DNA. We also rendered the human mtDNA into 2-dimensional images and compared it to the mtDNA of other species. On the search of the optimal image width we surprisingly detected a kind of fine grid within the mtDNA images which can be found in several species. The reason for the appearance of this grid is still unknown. If there is anyone who might have an explanation, she/he is cordially invited to comment it to us.
- When comparing the mtDNA of different species we wanted to get a deeper insight into their differences. This lead us to transcoding of not only the mtDNA itself but also of the amino acids of the proteins encoded by mtDNA.
Can the DNA sequence also be rendered in other ways?
- There are several ways to transcode data. For a visualization of the DNA sequence a simple form of transcoding is to give each of its bases a color and then generating images. The transcoding into an acoustical form (sonification) can be done by giving each base a voice instead of a color and streaming the sequence as a DNA-Radio. This idea is also quiet simple. The big challenge was how to prepare the huge amount of data for this project without crushing the server. (For comparison: A mp3 song of about 2-3 minutes has a size of about 2-3 Mb, but for streaming the sequence of e.g. the smallest of our chromosomes we need about 5,500 hours and about 49.5 Gb). The next step might be to give each base a tone or harmony to create some kind of interesting music.
What resources did you use?
- Besides reading countless articles from different authors, we have used the following sources that helped us a lot for creating this project: NCBI and the ideogram browser.
If the FAQ didn’t answer your question, please don’t hesitate to contact us.